Since the global eradication of smallpox in 1980, there has been little development of therapies for the variola virus. The global monkeypox outbreak in 2022 brought smallpox back into public focus, raising concerns that variola or monkeypox viruses could become potential bioweapons of terrorism. The variola virus and the monkeypox virus (MPXV) both belong to the Poxviridae family, Chordopoxvirinae subfamily, and Orthopoxvirus genus. Poxviruses are the largest known animal viruses, capable of infecting most vertebrates and invertebrates. They are double-stranded DNA viruses with about 200 distinct genes. A subset of these gene products can independently perform essential viral functions without the host cell nucleus, while others widely regulate the host cell and immune system.
Notable orthopoxviruses in human epidemiology include variola virus, vaccinia virus (VACV), cowpox virus (CPXV), and monkeypox virus (MPXV). Variola virus, the causative agent of smallpox, has two clinical epidemiological variants identified in the United States: Variola major virus and Variola minor virus (or alastrim virus), with fatality rates of 5-40% and 1%, respectively, indicating greater virulence of the Variola major virus. The cowpox virus was named for its association with pustular lesions on cow udders and milkmaids' hands. Interestingly, cows are not the natural host of CPXV, and cowpox is not a common disease. The vaccinia virus is widely used as a poxvirus model in laboratories and was used to produce smallpox vaccines, with strains like M-63 (Soviet Union), Lister (UK), Tiantan (China), and Wyeth, IHD (USA) being most commonly used.
The poxvirus genome is a linear double-stranded DNA with variable-length inverted terminal repeats forming hairpin structures. The full sequencing of vaccinia virus Copenhagen and WR strains shows a genome length of 191kbp, with 12kbp inverted terminal repeats (ITR) ending in covalently connected single-stranded hairpin loops. The sequence is AT-rich and contains short direct repeats and multiple ORFs. Poxviruses can replicate entirely in the cytoplasm, with low dependence on the host cell's DNA and RNA. The virus synthesizes new genes by generating and breaking down large tandem molecules, providing favorable conditions for genome amplification and evolution by acquiring new functions.
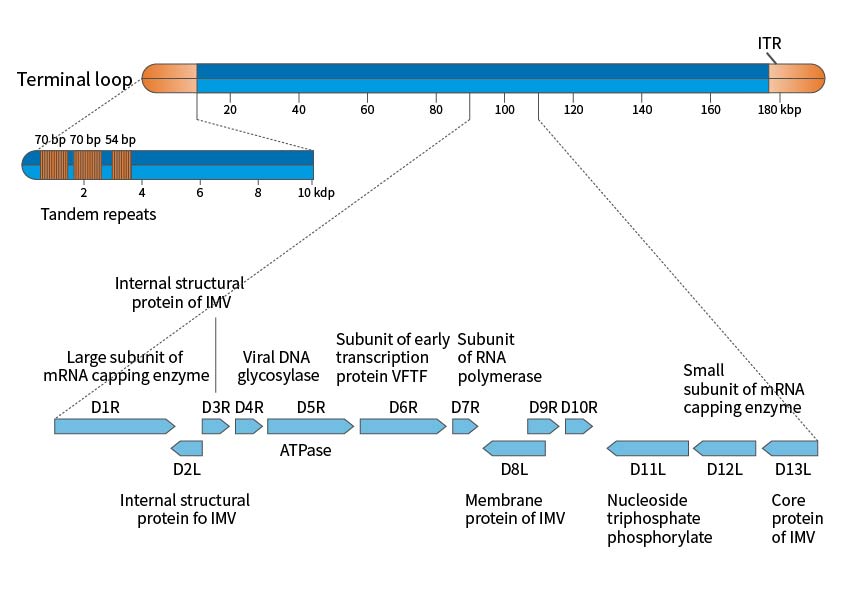
Figure 1. Schematic Diagram of the Vaccinia Virus Genome
Poxviruses are the most structurally complex known viruses, with elliptical or brick-like particles about 200-400nm long and an aspect ratio of 1.2-1.7. The membrane is a typical 50-55nm lipid-protein bilayer, surrounding the core and covered with randomly arranged tubular elements (STE), averaging 7nm wide and 100nm long. The virus particle consists of a core and related lateral bodies surrounded by a membrane, making it sufficiently infectious; however, for some virus strains and specific cell infections, the virus particle may acquire an additional lipid bilayer with a unique chemical composition, called the Envelope. This Envelope contains about twice the phospholipids of the non-enveloped virus particle. Studies have shown that antigens on the viral envelope can induce immunity, protecting the host from poxvirus infection. The poxvirus envelope contains at least seven different glycoproteins and a major non-glycosylated acylated polypeptide.
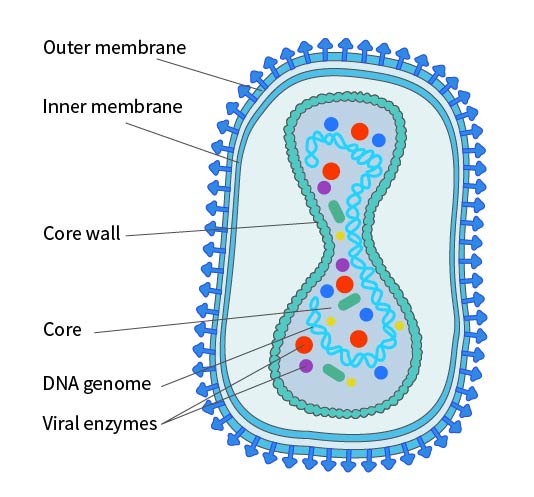
Figure 2. Structural Diagram of Poxvirus in IMV Form. Note: The existence of an inner membrane is still under debate.
The transmission process varies significantly depending on the poxvirus species and host, usually involving the following methods: 1) remaining free in the cytoplasm; 2) migrating to the cell surface and being expelled through microvilli; 3) being enveloped by a double membrane from the Golgi apparatus, transported to the cell surface, and released by an envelope (from the Golgi apparatus intraluminal pool membrane); 4) forming large vesicles through budding and acquiring an envelope; 5) combining with non-membrane vesicles; 6) combining with a protein substance in an acidophilic cytoplasmic structure called an a-type inclusion body (ATI).
The poxvirus replication cycle varies by virus species. Using the model virus VACV as an example, replication generally occurs 2-5 hours post-infection. Viral maturation occurs in five stages: immature virion (IV), intracellular mature virion (IMV), intracellular enveloped virion (IEV), cell-associated enveloped virion (CEV), and extracellular enveloped virion (EEV). IMV and EEV are the main infectious forms of vaccinia virus, with EEV being more infectious and entering cells through a non-pH-dependent membrane fusion process. EEV is responsible for intercellular transmission, eventually released as IMV into the external environment. Compared to EEV, IMV is more stable and responsible for inter-host transmission.
After fusion with the cell membrane, primary uncoating occurs, releasing the viral core into the cytoplasm. The core contains the viral genome, along with virus-dependent RNA polymerase, "initiation" proteins necessary for specific recognition of viral early gene promoters, and several RNA processing enzymes that modify viral transcripts. After release, the core synthesizes viral early mRNA (Step 2 in Figure 3), which exhibits the typical characteristics of cellular mRNA, being recognized and translated by the cell (Step 3 in Figure 3). About half of the viral genes are expressed early in infection. Some early proteins, similar in sequence to cellular growth factors, are secreted from the cell (Step 4 in Figure 3), inducing the proliferation of neighboring host cells or the suppression of host immune defense mechanisms. The synthesis of early proteins induces secondary uncoating, the core wall opens, and the nucleoprotein complex containing the genome is released from the core (Step 5 in Figure 3), halting the expression of viral early genes.
Early proteins catalyze the replication of the viral DNA genome (Step 6 in Figure 3), and the newly synthesized viral DNA molecules serve as templates for the next replication cycle (Step 7 in Figure 3) and as templates for the transcription of viral intermediate genes (Step 8 in Figure 3). Intermediate gene transcription requires specific viral proteins (products of early genes) that confer specificity for intermediate promoters on the viral RNA polymerase and host cell proteins (Vitf2) relocated from the infected cell nucleus to the cytoplasm. Intermediate mRNA encodes proteins (Step 9 in Figure 3), including those needed for late gene transcription (Step 10 in Figure 3). Late genes encode proteins that construct the virus particle, enzymes, and other early initiation proteins needed for virus particle assembly. These viral proteins are synthesized with the help of the cellular translation mechanism (Step 11 in Figure 3).
Viral membrane proteins are non-glycosylated, and the role of the cellular membrane in early assembly is controversial (Step 12 in Figure 3). Initial assembly forms immature virus particles (Step 13 in Figure 3), spherical particles separated by a membrane, possibly obtained from the early secretion pathway of the cell. The virus particle matures into brick-shaped IMV (Step 14 in Figure 3), released upon cell lysis (Step 15 in Figure 3). Additionally, the particles can acquire a second bilayer from the Golgi apparatus or early endosome, forming intracellular enveloped virus particles (IEV) (Step 16 in Figure 3).
IEV moves to the cell surface on microtubules, fusing with the plasma membrane to form cell-associated virus particles (CEV) (Step 17 in Figure 3). These CEV induce actin polymerization, promoting the direct transfer of actin to surrounding cells (Step 18 in Figure 3); they can also separate from the membrane to form EEV.
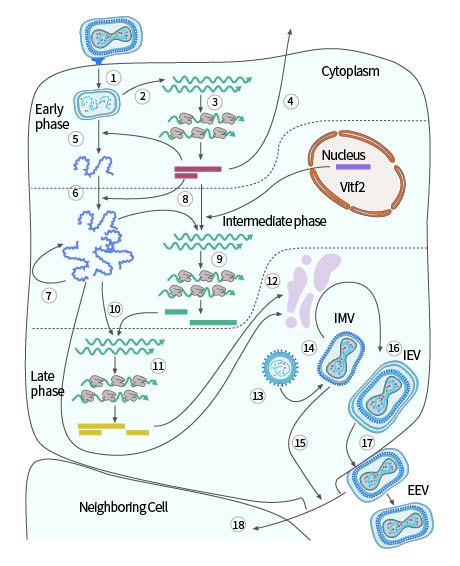
Figure 3. Single-cell reproduction cycle of vaccinia virus
References
1.Buller RM, Palumbo GJ. Poxvirus pathogenesis. Microbiol Rev. 1991, 55(1): 80-122.
2.Harrison SC, Alberts B, Ehrenfeld E, et al. Discovery of antivirals against smallpox. Proc Natl Acad Sci U S A. 2004, 101(31): 11178-11192.
