Monkeypox virus (MPV) belongs to the Orthopoxvirus genus of the Poxviridae family and causes a human disease similar to smallpox. The 196,858 bp genome of MPV was analyzed for structural features and open reading frames (ORFs). Each end of the genome contains a terminal inverted repeat (ITR) sequence that is 6379 bp long, completely identical but oriented in the opposite direction, a putative telomere resolution sequence, and short tandem repeat sequences. MPV contains the core genes of known Orthopoxviruses but only a subset of the immune modulation and host range genes. Sequence comparison confirms that MPV is a distinct Orthopoxvirus, neither a direct ancestor nor a direct descendant of the Variola virus.
Poxviridae comprises complex double-stranded DNA viruses that replicate in the cytoplasm of vertebrate or invertebrate cells. Orthopoxviruses include Variola virus (VAR), Monkeypox virus (MPV), Cowpox virus (CPV), and Vaccinia virus (VAC). VAR is the causative agent of human smallpox, a disease with a mortality rate of 10%-40%, eradicated through a strategic live VAC vaccination campaign. After smallpox eradication, vaccination ceased, leading to decreased immunity against other Orthopoxviruses. Consequently, susceptibility to zoonotic viruses such as MPV, CPV, and VAC has increased. These viruses' potential adaptation to increased pathogenicity or transmissibility in human populations could occur.
The similar clinical presentation of monkeypox and smallpox led to the hypothesis that MPV is an evolutionary ancestor of VAR. MPV and VAR evolution was analyzed based on genome restriction endonuclease maps or nucleotide sequence comparisons of single viral genes. The complete genome of human MPV isolate ZAI-96-I-16 (MPV-ZAI) was sequenced. Whole-genome comparison concluded that MPV is neither a direct ancestor nor a descendant of VAR. Detailed analysis of the MPV-ZAI DNA sequence, compared with corresponding whole-genome sequences of VAC, VAR, and partial CPV sequences, showed that the sequence includes the entire genome except for part of the covalent terminal hairpin loop.
identical but oriented in the opposite direction, known as the terminal inverted repeat (ITR), which includes a set of short tandem repeat sequences and terminal hairpins. The MPV-ZAI genome contains a 6379 bp ITR. One of the two complementary, incompletely base-paired hairpin loop strands in MPV-ZAI DNA was successfully cloned through S1 nuclease digestion and DNA polymerase I polymerization. This sequence, aligned with other Orthopoxvirus sequences, showed considerable conservation, with only four nucleotides missing from the loop-forming region (Figure 1).

Figure 1. Comparison of the terminal regions of MPV and other Orthopoxviruses
Note: The upper line shows the ordered portion of the terminal loop of MPV-ZAI. Nucleotides differ from the VAC-WR sequence. ZAI, BSH, and WR represent Zaire-96-I-16, Bangladesh, and West Reserve virus strains, respectively.
The telomere resolution sequence of MPV is identical to those of VAC and VAR. The tandem repeat sequence region near the terminal hairpin of the MPV-ZAI genome is shorter, consisting of NR I (85 bp) and NR II (322 bp). The organization of this region in MPV-ZAI DNA is most similar to VAR-GAR (Figure 2). However, the terminal regions of all VAR strains have very short ITRs, lacking ORFs, and differing sets of tandem repeat sequences at the left and right ends.
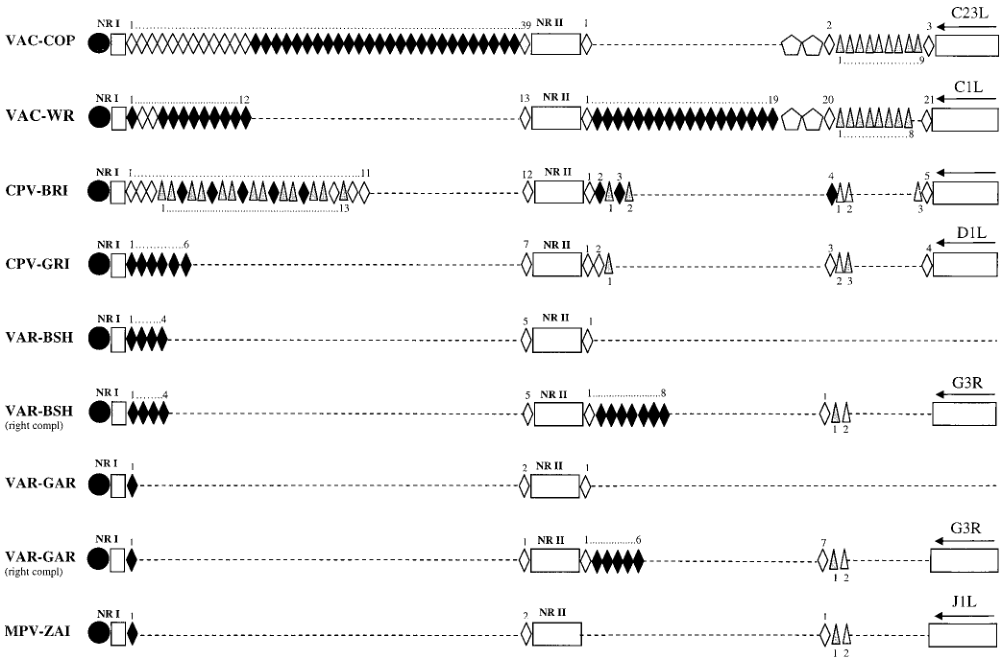
Figure 2. Tandem repeat pattern within the ITR region of MPV and other Orthopoxviruses
Note: The white rectangles represent unique ITR sequences: NR I, NR II, and coding regions. Black diamonds correspond to 70 bp tandem repeat sequences, with the numbers indicated above. Gray triangles correspond to 54 bp repeats, with the numbers shown below. Pentagons in VAC-COP and VAC-WR ITR sequences represent 125 bp repeats. Open squares and triangles indicate variations from the consensus sequence through deletions, replacements, or insertions of these repeats. Black circles represent terminal hairpins. Dashed lines indicate the absence of corresponding DNA.
Four ORFs on the left side of the MPV-ZAI genome (Figure 3A) are located within the ITR, so corresponding ORFs also exist on the right side of the genome (Figure 3B). All genes known to be crucial in other Orthopoxviruses are present in MPV and occupy the central region of the genome (ORFs C10L to A25R). These ORFs share more than 90% sequence homology with those of other Orthopoxviruses. Most species and strain-specific differences between MPV-ZAI and other Orthopoxviruses are found in the terminal regions at the left and right ends (Figures 3A and 3B). MPV-ZAI lacks 25 ORFs found in CPV-GRI and 19 potential ORFs in VAR-IND. Although the functions of some of these genes remain unknown, they may include many involved in immune evasion, host range, and cell proliferation.
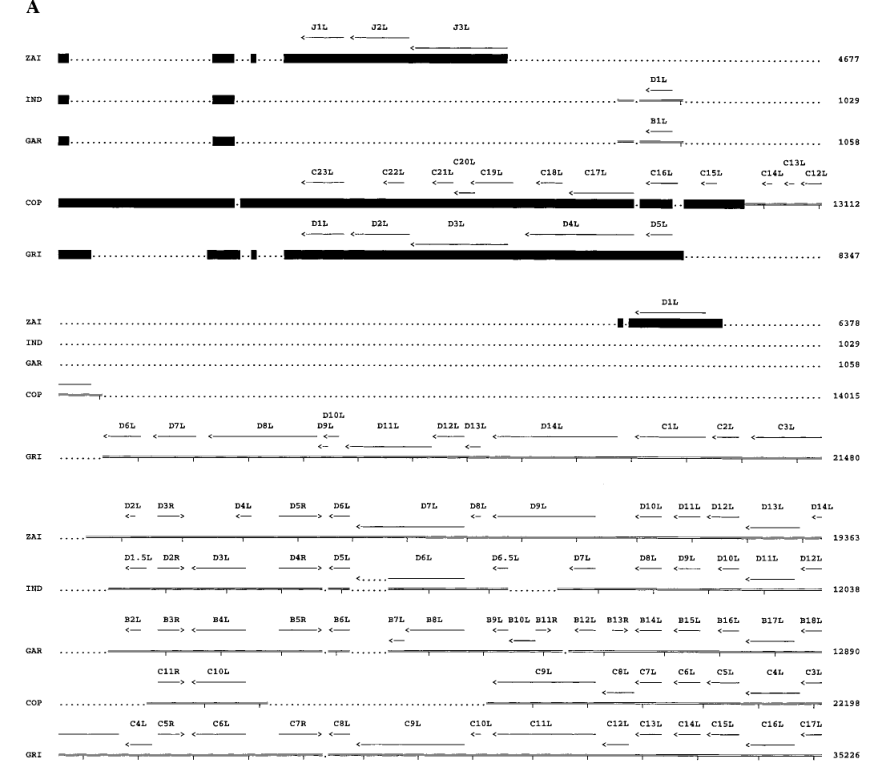

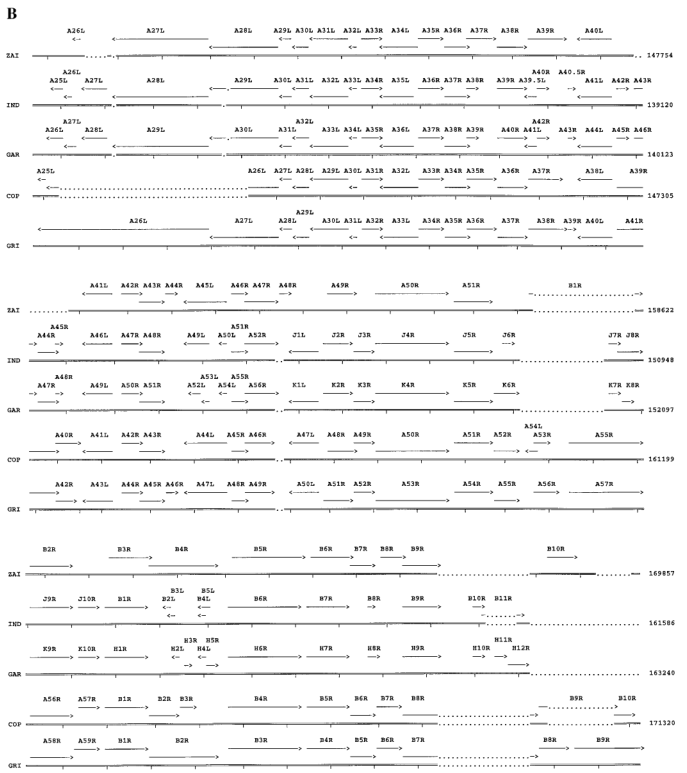
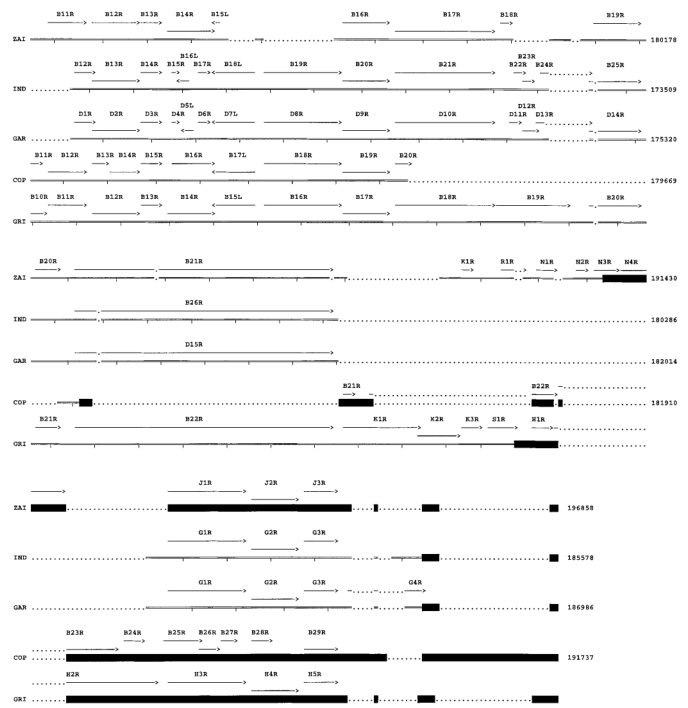
Figure 3. Comparison of ORFs in the left (A) and right (B) terminal variable regions of MPV with corresponding genomic fragments of other Orthopoxviruses.
Note: ORF sizes and directions are indicated by arrows. Deletions of DNA and protein relative to another virus, exceeding 150 bp and 50 amino acids, respectively, are marked with dots. Black squares mark the ITR region. Nucleotide numbering is shown on the right.
Previous comparisons of MPV and VAR genes added VAC and CPV growth factors and immune evasion genes in Table 1, encompassing all such known genes present in Orthopoxviruses. Based on the number of intact and broadly truncated virulence genes (indicated in parentheses), the following order was derived: CPV-GRI (17) > VAR-IND (11) = VAR-GAR (11) > MPV-ZAI (10) > VAC-COP (9) > VAC-MVA (6). Therefore, MPV-ZAI also contains fewer ankyrin repeat genes than CPV.
Table 1. Orthopoxvirus Growth Factors and Immune Evasion Genes
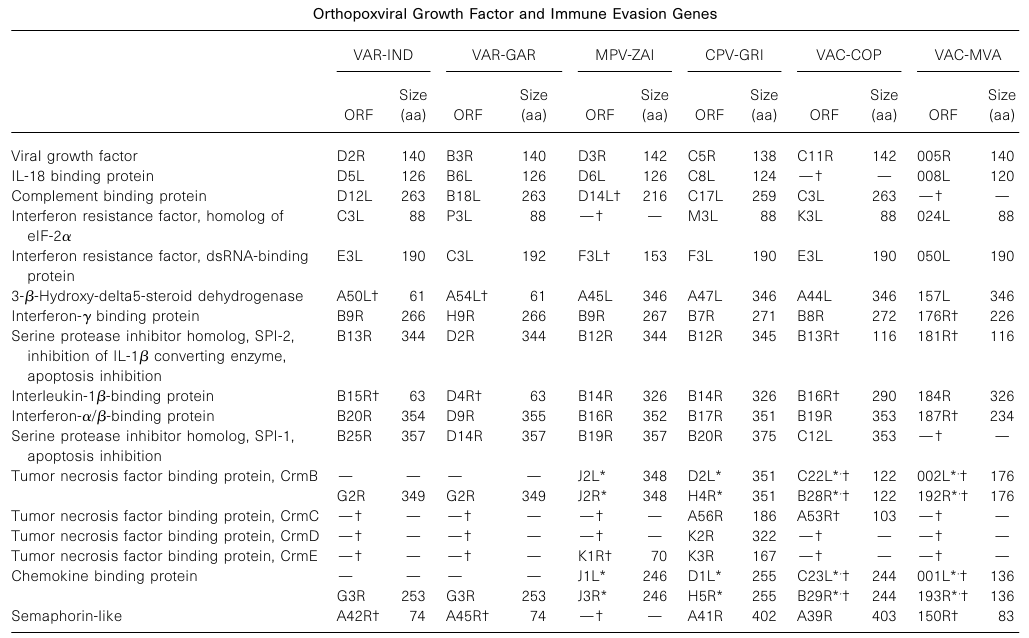
Note: * indicates ORFs duplicated in the inverted terminal repeat (ITR) regions on the left and right sides. † indicates ORFs with significant functional differences from corresponding ORFs in CPV-GRI.
CPV-GRI encodes the Kelch family 6-500 amino acids of unknown function, sharing 22-26% similarity with each other. While VAC-COP encodes three Kelch family proteins with homology to CPV-GRI proteins at 99.4%, 97.9%, and 98.6%, respectively, the MPV-ZAI genome encodes only one C9L, homologous to CPV-GRI protein G3L with 97.3% similarity. In the VAR genome, these ORFs appear disrupted, suggesting they are not essential for replication.
VAC contains two ORFs with phospholipase D sequences: F13L is necessary for the formation of extracellular virions and effective intercellular spread of infection, while K4L is essential for replication in tissue culture. MPV-ZAI contains intact versions of these three ORFs (C4L, C5L, and C19L), whereas VAR strains lack C4L and C5L homologs due to DNA fragment deletions (Figure 3A). The functions of C4L and C5L proteins are unclear, but they are encoded by both MPV and CPV, two Orthopoxviruses with relatively broad host ranges.
Based on the DNA sequence of the terminal variable region, it is believed that the organizational structure of CPV is most similar to the ancestral orthopoxvirus species, and the distance between VAR and MPV is slightly farther than that of VAC. However, by analyzing the left and right variable regions separately, the situation appears to be more complicated. VAC strains appeared to be closer to the MPV-ZAI terminal variable region than their left side and VAR strains relative to the right terminal variable region (Figure 4), this difference may be due to the complex recombination origin of VAC.
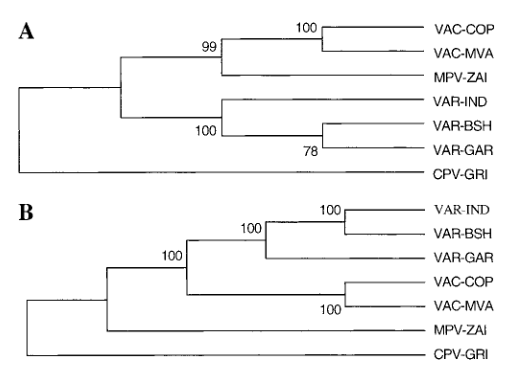
Figure 4. Phylogenetic analysis
The monkeypox virus (MPV) genome shows a distinct structure compared to other Orthopoxviruses, such as variola (VAR), cowpox (CPV), and vaccinia (VAC) viruses. The MPV genome is neither an ancestor nor a direct descendant of the VAR genome, suggesting independent evolutionary paths. The analysis also reveals differences in host range and immune evasion genes among Orthopoxviruses, with MPV having fewer such genes than CPV but more than VAR. These findings contribute to understanding MPV's evolutionary history and its relationship with other Orthopoxviruses.
References
1. Shchelkunov SN, Totmenin AV, Safronov PF, et al. Analysis of the monkeypox virus genome[J]. Virology. 2002, 297(2): 172-194.
